The Dataset
Our goal for this experiment is to determine which Arabidopsis thaliana genes respond to nitrate. The dataset is a simple experiment where RNA is extracted from roots of independent plants and then sequenced. Two plants were treated with the control (KCl) and two samples were treated with Nitrate (KNO3).
The sequences were processed to remove all low quality sequences, trim all low quality nucleotides, and finally aligned against the Arabidopsis genome using TopHat. Our analysis starts from here.
We have been provided the following files:
- 4 Bam files – An alignment file, one for each sample
- Arabidopsis.gtf file – which contains information about the genes in Arabidopsis and where they are located in the genome.
- Experiment design – a comma separated file containing meta data.
- Gene description – Description about the function of the genes in Arabidopsis.
If you are doing this for the first time, you will have to install some packages first.
If you are on Dalma, please see special instructions on how to install the packages below.
##########################################################################################
# Note: If one of the R packages is not loaded, you have to install it from Bioconductor
# Using the code below. It is a good idea to make sure if it's already installed by
# first typing :
# library()
##########################################################################################
source("http://bioconductor.org/biocLite.R")
biocLite("Rsamtools")
biocLite("GenomicFeatures")
biocLite("GenomicAlignments")
biocLite("DESeq2")
biocLite("GOstats")
biocLite("GO.db")
biocLite("Category")
biocLite("org.At.tair.db")
The Data Files
We will use the result from the previous Aligning RNA-seq section.
It’s a good idea to start R from within the directory where the files are located.
BAM files are provided in this folder just in case you are not able to finish the alignment in time
cd $SCRATCH/rnaseq_alignment/
Running R in interactive mode and modules to load.
module load gencore module load gencore_build module load gencore_base module load gencore_rnaseq srun --mem 8GB --time 12:00:00 --pty /bin/bash # to start R, just type R R
###
The Alignment Files
The alignment files are in bam format. This files will not be loaded into R, but rather simply pointed to by a reference/variable. The alignment files provided are about 15x smaller compared to an average RNA-seq sample run today. Also there will be triplicates of 3 or more different conditions resulting in much more than 4 sample. So you can imagine the amount of space and memory R would need if all that data was stored in the workspace.
To do point to the bam files we will use Rsamtools
library(Rsamtools)
Loading required package: GenomeInfoDb
Loading required package: BiocGenerics
Loading required package: parallel
Attaching package: ‘BiocGenerics’
The following objects are masked from ‘package:parallel’:
clusterApply, clusterApplyLB,
clusterCall, clusterEvalQ,
clusterExport, clusterMap,
parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply,
parSapplyLB
The following objects are masked from ‘package:stats’:
IQR, mad, sd, var, xtabs
The following objects are masked from ‘package:base’:
anyDuplicated, append,
as.data.frame, cbind, colMeans,
colnames, colSums, do.call,
duplicated, eval, evalq, Filter,
Find, get, grep, grepl, intersect,
is.unsorted, lapply, lengths, Map,
mapply, match, mget, order, paste,
pmax, pmax.int, pmin, pmin.int,
Position, rank, rbind, Reduce,
rowMeans, rownames, rowSums,
sapply, setdiff, sort, table,
tapply, union, unique, unsplit,
which, which.max, which.min
Loading required package: S4Vectors
Loading required package: stats4
Attaching package: ‘S4Vectors’
The following object is masked from ‘package:base’:
expand.grid
Loading required package: IRanges
Loading required package: GenomicRanges
Loading required package: Biostrings
Loading required package: XVector
Attaching package: ‘Biostrings’
The following object is masked from ‘package:base’:
strsplit
(bamfiles = BamFileList(dir(pattern="dedup.bam")))
## BamFileList of length 4 ## names(4): KCL_1_dedup.bam KCL_2_dedup.bam NO3_1_dedup.bam NO3_2_dedup.bam
The Annotation File
GTF file is very similar to a GFF file. Their purpose is to store the location of genome features, such as genes, exons, and CDS. It is a tab delimited flat file which looks something like this.
## reference source feature start stop score strand phase
## 1 Chr1 TAIR10 exon 3631 3913 . + .
## 2 Chr1 TAIR10 exon 3996 4276 . + .
## 3 Chr1 TAIR10 exon 4486 4605 . + .
## 4 Chr1 TAIR10 exon 4706 5095 . + .
## 5 Chr1 TAIR10 exon 5174 5326 . + .
## 6 Chr1 TAIR10 exon 5439 5899 . + .
## annotation
## 1 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
## 2 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
## 3 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
## 4 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
## 5 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
## 6 transcript_id AT1G01010.1; gene_id AT1G01010; gene_name AT1G01010;
We will load the GTF file using GenomicFeatures and group the exons based on the gene annotation.
library(GenomicFeatures)
## Loading required package: AnnotationDbi
## Loading required package: Biobase
## Welcome to Bioconductor
##
## Vignettes contain introductory
## material; view with
## 'browseVignettes()'. To cite
## Bioconductor, see
## 'citation("Biobase")', and for
## packages 'citation("pkgname")'.
txdb = makeTxDbFromGFF("Arabidopsis.gtf", format="gtf")
## Import genomic features from the file as a GRanges object ... OK
## Prepare the 'metadata' data frame ... OK
## Make the TxDb object ... OK
(ebg = exonsBy(txdb, by="gene"))
## GRangesList object of length 33610:
## $AT1G01010
## GRanges object with 6 ranges and 2 metadata columns:
## seqnames ranges strand | exon_id exon_name
## |
## [1] Chr1 [3631, 3913] + | 1
## [2] Chr1 [3996, 4276] + | 2
## [3] Chr1 [4486, 4605] + | 3
## [4] Chr1 [4706, 5095] + | 4
## [5] Chr1 [5174, 5326] + | 5
## [6] Chr1 [5439, 5899] + | 6
##
## $AT1G01020
## GRanges object with 12 ranges and 2 metadata columns:
## seqnames ranges strand | exon_id exon_name
## [1] Chr1 [5928, 6263] - | 21880
## [2] Chr1 [6437, 7069] - | 21881
## [3] Chr1 [6790, 7069] - | 21882
## [4] Chr1 [7157, 7232] - | 21883
## [5] Chr1 [7157, 7450] - | 21884
## ... ... ... ... ... ... ...
## [8] Chr1 [7762, 7835] - | 21887
## [9] Chr1 [7942, 7987] - | 21888
## [10] Chr1 [8236, 8325] - | 21889
## [11] Chr1 [8417, 8464] - | 21890
## [12] Chr1 [8571, 8737] - | 21891
##
## $AT1G01030
## GRanges object with 2 ranges and 2 metadata columns:
## seqnames ranges strand | exon_id exon_name
## [1] Chr1 [11649, 13173] - | 21892
## [2] Chr1 [13335, 13714] - | 21893
##
## ...
## <33607 more elements>
## -------
## seqinfo: 7 sequences (2 circular) from an unspecified genome; no seqlengths
The Experimental Design
We will need meta information about our samples to define which sample is the KCL and which is the KNO3. A simple comma separated file can be created using a text editor where the first column has the sample file names and the second column has the categories for the samples. The file should look something like this:

Now let’s load the file into our workspace. The commands read.table, read.csv, and read.delim are all related with the exception of different default parameters. They all read a text file and load it into your workspace as a data.frame. Additonally it loads all columns that are character vectors and makes them factors. The first option for all the different versions is the neame of the file. After that, unless you have memorized the options for the commands, explicitly state which parameters you are providing.
expdesign = read.csv("expdesign.txt", row.names=1, sep=",")
Counting the Reads
Now we will use the GenomicAlignments package to determine where the reads are mapping. Remember we decided to map the sequences to the genome so we can make new discoveries such as novel genes, alternative splicing, and also anti-sense transcripts. In the event that the reference genome is not sequenced, one would have to assemble the RNA-seq reads first to identify all the genes that were detected in the RNA-seq samples. However assembling the transcriptome is quite a complicated process and requires a lot of time and manual curation to produce quality transcripts. We are fortunate that we are using Arabidopsis data whose genome was sequenced in 2000.
The function summarizeOverlaps takes the bam files and the gene annotation to count the number of reads that are matching a gene. The union mode means if a read matches an area where two genes overlap, then it is not counted. We can also provide other parameters such as whether sequence was single-end or pair-end.
library("GenomicAlignments")
## Loading required package: SummarizedExperiment
## Loading required package: DelayedArray
## Loading required package: matrixStats
##
## Attaching package: ‘matrixStats’
##
## The following objects are masked from ‘package:Biobase’:
##
## anyMissing, rowMedians
##
##
## Attaching package: ‘DelayedArray’
##
## The following objects are masked from ‘package:matrixStats’:
##
## colMaxs, colMins, colRanges,
## rowMaxs, rowMins, rowRanges
##
## The following object is masked from ‘package:Biostrings’:
##
## type
##
## The following object is masked from ‘package:base’:
##
## apply
se = summarizeOverlaps(features=ebg,
reads=bamfiles, mode="Union", singleEnd=TRUE,
ignore.strand=TRUE )
counts = assay(se)
countgenenames = gsub("[.][1234567890]", "", row.names(counts))
rownames(counts)=countgenenames
Filtering the Counts
Now, if you remember from the lecture, genes that are expressed at a very low level are extremely unreliable. We will remove all genes if neither of the groups ( KCL or KNO3 ) have a median count of 10 and call the new dataframe counts_filtered.
medianCountByGroup = t(apply(counts, 1, tapply,
expdesign, median))
maxMedian=apply(medianCountByGroup, 1, max)
counts_filtered = counts[maxMedian >= 10,]
Differentially Expressed Genes
Now that we have the counts table filtered, we can start to determine if any of the genes are significantly differentially expressed using DESeq. DESeq performs a pairwise differential expression test by creating a negative binomial model.
Now we can create an object that DESeq needs using the function newCountDataSet. In order to create this dataset, we need the filtered data frame of read counts and the factor that will help group the data based on the condition.
library(DESeq2)
cds = DESeqDataSetFromMatrix(countData=counts_filtered, colData=expdesign, design= ~ condition) # if you would like to try to run without the filtering # simply commend the above lines and uncomment below. #cds = DESeqDataSetFromMatrix(countData=counts, # colData=expdesign, # design= ~ condition)
Before the actual test, DESeq has to consider the difference in total reads from the different samples. This is done by using estimateSizeFactors function.
cds = estimateSizeFactors(cds)
Next DESeq will estimate the dispersion ( or variation ) of the data. If there are no replicates, DESeq can manage to create a theoretical dispersion but this is not ideal.
cds = estimateDispersions( cds )
## gene-wise dispersion estimates
## mean-dispersion relationship
## final dispersion estimates
plotDispEsts( cds )
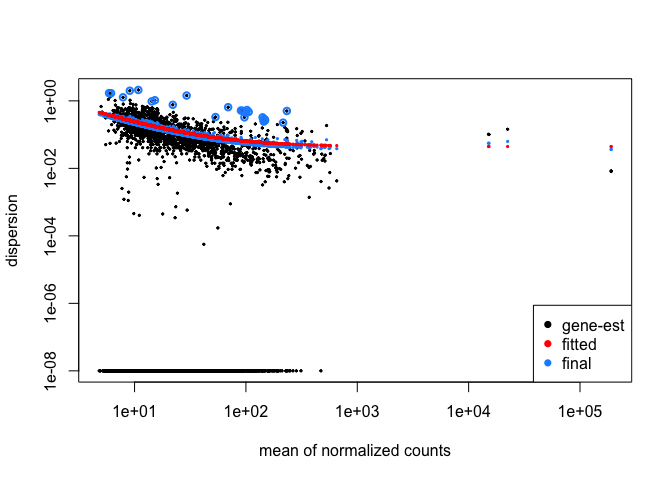
The plot shows us that as the gene’s read count increases, dispersion decreases, which is what we expect. Now we will tell DESeq what we would like to compare. Then we will use the adjusted p-value ( p-value corrected for multiple hypothesis testing ) for our cutoff.
cds <- DESeq(cds)
## using pre-existing size factors
## estimating dispersions
## found already estimated dispersions, replacing these
## gene-wise dispersion estimates
## mean-dispersion relationship
## final dispersion estimates
## fitting model and testing
res <- results(cds) head(res)
## log2 fold change (MAP): condition treated vs control
## Wald test p-value: condition treated vs control
## DataFrame with 6 rows and 6 columns
## baseMean log2FoldChange lfcSE stat pvalue
##
## AT1G01010 12.32304 -0.16338576 0.4536306 -0.36017355 0.7187174
## AT1G01040 19.94966 -0.18960085 0.4240140 -0.44715708 0.6547616
## AT1G01050 11.53394 0.30014338 0.4569963 0.65677416 0.5113261
## AT1G01060 26.21540 -0.02992785 0.4110043 -0.07281639 0.9419522
## AT1G01090 34.07506 0.19051694 0.3836878 0.49654152 0.6195124
## AT1G01100 122.42019 0.36447662 0.3037174 1.20005195 0.2301192
## padj
##
## AT1G01010 0.9995993
## AT1G01040 0.9995993
## AT1G01050 0.9995993
## AT1G01060 0.9995993
## AT1G01090 0.9995993
## AT1G01100 0.9995993
sum(res$padj < 0.05, na.rm=T)
## [1] 91
The sum command is normally used to add numberic values. However in logic vectors, TRUE=1 and FALSE=0, so we can use the sum function to count the number of “TRUE” in the vector. In the example we counted the number of genes that have a an adjusted p-value less than 0.05.
MA Plot
Here’s an MA plot that shows as the average count of a gene increases, a smaller fold change is needed for something to be significant. For this reason, it is often helpful to require that the log2foldchange also be greater than or less than negative of some cutoff.
plotMA(res, ylim=c(-5,5))
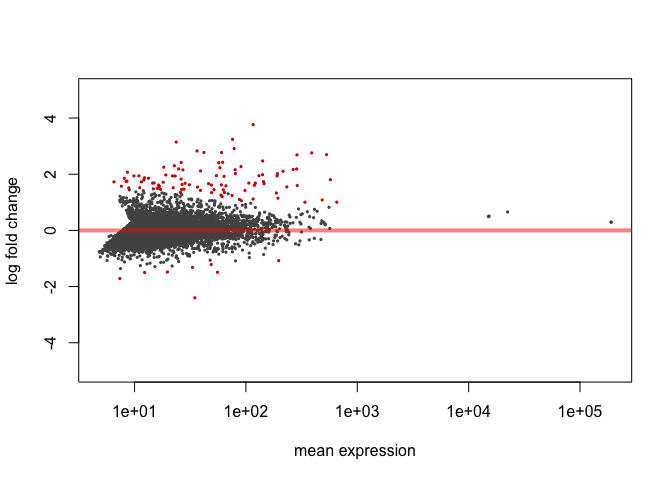
Significant genes
Let’s use the same values for our cutoff to determine which genes we want to consider as significantly differentially expressed. The resSigind variable will contain genes that are induced and resSigrep will contain genes that are repressed when the plants are treated with Nitrate. To create one dataframe of differentially expressed genes, let’s combine the two dataframe. We can use the rbind command because the columns are the same in both sets. To show the name of the genes, simply look in the id column of the dataframe.
resSigind = res[ which(res$padj < 0.05 & res$log2FoldChange > 0), ] resSigrep = res[ which(res$padj < 0.05 & res$log2FoldChange < 0), ] resSig = rbind(resSigind, resSigrep) rownames(resSigind)
## [1] "AT1G07150" "AT1G12110" "AT1G22170" "AT1G22500" "AT1G23870"
## [6] "AT1G24280" "AT1G25550" "AT1G30510" "AT1G35560" "AT1G43710"
## [11] "AT1G49860" "AT1G61740" "AT1G62180" "AT1G63940" "AT1G64190"
## [16] "AT1G70410" "AT1G74030" "AT1G77760" "AT1G78050" "AT1G80380"
## [21] "AT2G01140" "AT2G15620" "AT2G16060" "AT2G26980" "AT2G27510"
## [26] "AT2G28780" "AT2G30670" "AT2G33550" "AT2G36580" "AT2G41560"
## [31] "AT2G48080" "AT3G07350" "AT3G18560" "AT3G27170" "AT3G28510"
## [36] "AT3G30415" "AT3G31161" "AT3G47520" "AT3G47980" "AT3G48990"
## [41] "AT3G49940" "AT3G52360" "AT3G52930" "AT3G52960" "AT3G58610"
## [46] "AT3G60750" "AT4G02380" "AT4G05390" "AT4G09620" "AT4G17550"
## [51] "AT4G24620" "AT4G24670" "AT4G26970" "AT4G29905" "AT4G30190"
## [56] "AT4G31730" "AT4G31910" "AT4G32950" "AT4G34860" "AT4G35260"
## [61] "AT4G36010" "AT5G03330" "AT5G03380" "AT5G04950" "AT5G09800"
## [66] "AT5G10030" "AT5G10210" "AT5G12860" "AT5G13110" "AT5G13420"
## [71] "AT5G14760" "AT5G19120" "AT5G19970" "AT5G35630" "AT5G40850"
## [76] "AT5G41670" "AT5G49480" "AT5G50200" "AT5G51830" "AT5G53460"
## [81] "AT5G54170" "AT5G58320" "AT5G63160" "AT5G65030" "AT5G65210"
## [86] "AT5G67420"
rownames(resSigrep)
## [1] "AT1G47128" "AT1G64590" "AT1G67810" "AT4G17670" "AT5G16570"
Gene Annotations
Great ! We have genes that are differentially expressed, what do we know about these genes ? The gene identifier we obtained from the GTF file is referred to as TAIR identifiers (a consortium that used to release Arabidopsis genome annotations) I managed to download the gene description for all the genes. Let’s load them into the workspace and find out what are the names of the genes.Since the set of repressed genes is smaller, let’s see what we can find out about them.
Before we do this, note that the identified in the gene_description file is slightly different ( the file contains the transcript identifier that ends with “.” and a number), Let’s replace every occurrence of . and a number with nothing. Then we will be able to use match to find where our gene is in the description file so we can only print out that row.
gene_description <- read.delim("gene_description_20131231.txt",
header=FALSE, quote="")
genenames = gsub("[.][1234567890]", "",
gene_description[,1])
gene_description[,1]=genenames
gene_match_rows=match(rownames(resSig), gene_description[,1])
gene_description[gene_match_rows,c(1,3)]
## V1
## 376 AT1G07150
## 23961 AT1G12110
## 1354 AT1G22170
## 648 AT1G22500
## 21277 AT1G23870
## 3197 AT1G24280
## 3017 AT1G25550
## 2651 AT1G30510
## 1306 AT1G35560
## 3004 AT1G43710
## 371 AT1G49860
## 20768 AT1G61740
## 1652 AT1G62180
## 20722 AT1G63940
## 2156 AT1G64190
## 1486 AT1G70410
## 3058 AT1G74030
## 21700 AT1G77760
## 2778 AT1G78050
## 3462 AT1G80380
## 4049 AT2G01140
## 5984 AT2G15620
## 5894 AT2G16060
## 6762 AT2G26980
## 4037 AT2G27510
## 5918 AT2G28780
## 6094 AT2G30670
## 5620 AT2G33550
## 4915 AT2G36580
## 7274 AT2G41560
## 7813 AT2G48080
## 8618 AT3G07350
## 9735 AT3G18560
## 11045 AT3G27170
## 11579 AT3G28510
## 22970 AT3G30415
## 23117 AT3G31161
## 8534 AT3G47520
## 11468 AT3G47980
## 11986 AT3G48990
## 9111 AT3G49940
## 11870 AT3G52360
## 9447 AT3G52930
## 9455 AT3G52960
## 8137 AT3G58610
## 12062 AT3G60750
## 14687 AT4G02380
## 12545 AT4G05390
## 15120 AT4G09620
## 14117 AT4G17550
## 13247 AT4G24620
## 13258 AT4G24670
## 12586 AT4G26970
## 28092 AT4G29905
## 14080 AT4G30190
## 13579 AT4G31730
## 12645 AT4G31910
## 13452 AT4G32950
## 12660 AT4G34860
## 13264 AT4G35260
## 14907 AT4G36010
## 15719 AT5G03330
## 15710 AT5G03380
## 19440 AT5G04950
## 15942 AT5G09800
## 20427 AT5G10030
## 25683 AT5G10210
## 20234 AT5G12860
## 19953 AT5G13110
## 20165 AT5G13420
## 20570 AT5G14760
## 20226 AT5G19120
## 16192 AT5G19970
## 18367 AT5G35630
## 18205 AT5G40850
## 17697 AT5G41670
## 25085 AT5G49480
## 17377 AT5G50200
## 18302 AT5G51830
## 19786 AT5G53460
## 16883 AT5G54170
## 17817 AT5G58320
## 17909 AT5G63160
## 19701 AT5G65030
## 19024 AT5G65210
## 17435 AT5G67420
## 22519 AT1G47128
## 1820 AT1G64590
## 22251 AT1G67810
## 14147 AT4G17670
## 27313 AT5G16570
## V3
## 376 mitogen-activated protein kinase kinase kinase 13
## 23961 nitrate transporter 1.1
## 1354 Phosphoglycerate mutase family protein
## 648 RING/U-box superfamily protein
## 21277 trehalose-phosphatase/synthase 9
## 3197 glucose-6-phosphate dehydrogenase 3
## 3017 myb-like transcription factor family protein
## 2651 root FNR 2
## 1306 TCP family transcription factor
## 3004 Pyridoxal phosphate (PLP)-dependent transferases superfamily protein
## 371 glutathione S-transferase (class phi) 14
## 20768 Sulfite exporter TauE/SafE family protein
## 1652 5'adenylylphosphosulfate reductase 2
## 20722 monodehydroascorbate reductase 6
## 2156 6-phosphogluconate dehydrogenase family protein
## 1486 beta carbonic anhydrase 4
## 3058 enolase 1
## 21700 nitrate reductase 1
## 2778 phosphoglycerate/bisphosphoglycerate mutase
## 3462 P-loop containing nucleoside triphosphate hydrolases superfamily protein
## 4049 Aldolase superfamily protein
## 5984 nitrite reductase 1
## 5894 hemoglobin 1
## 6762 CBL-interacting protein kinase 3
## 4037 ferredoxin 3
## 5918
## 6094 NAD(P)-binding Rossmann-fold superfamily protein
## 5620 Homeodomain-like superfamily protein
## 4915 Pyruvate kinase family protein
## 7274 autoinhibited Ca(2+)-ATPase, isoform 4
## 7813 oxidoreductase, 2OG-Fe(II) oxygenase family protein
## 8618 Protein of unknown function (DUF506)
## 9735
## 11045 chloride channel B
## 11579 P-loop containing nucleoside triphosphate hydrolases superfamily protein
## 22970
## 23117
## 8534 malate dehydrogenase
## 11468 Integral membrane HPP family protein
## 11986 AMP-dependent synthetase and ligase family protein
## 9111 LOB domain-containing protein 38
## 11870
## 9447 Aldolase superfamily protein
## 9455 Thioredoxin superfamily protein
## 8137 ketol-acid reductoisomerase
## 12062 Transketolase
## 14687 senescence-associated gene 21
## 12545 root FNR 1
## 15120 Mitochondrial transcription termination factor family protein
## 14117 Major facilitator superfamily protein
## 13247 phosphoglucose isomerase 1
## 13258 tryptophan aminotransferase related 2
## 12586 aconitase 2
## 28092
## 14080 H(+)-ATPase 2
## 13579 glutamine dumper 1
## 12645 HXXXD-type acyl-transferase family protein
## 13452 Protein phosphatase 2C family protein
## 12660 Plant neutral invertase family protein
## 13264 isocitrate dehydrogenase 1
## 14907 Pathogenesis-related thaumatin superfamily protein
## 15719 Cysteine proteinases superfamily protein
## 15710 Heavy metal transport/detoxification superfamily protein
## 19440 nicotianamine synthase 1
## 15942 ARM repeat superfamily protein
## 20427 TGACG motif-binding factor 4
## 25683
## 20234 dicarboxylate transporter 1
## 19953 glucose-6-phosphate dehydrogenase 2
## 20165 Aldolase-type TIM barrel family protein
## 20570 L-aspartate oxidase
## 20226 Eukaryotic aspartyl protease family protein
## 16192
## 18367 glutamine synthetase 2
## 18205 urophorphyrin methylase 1
## 17697 6-phosphogluconate dehydrogenase family protein
## 25085 Ca2+-binding protein 1
## 17377 nitrate transmembrane transporters
## 18302 pfkB-like carbohydrate kinase family protein
## 19786 NADH-dependent glutamate synthase 1
## 16883 Polyketide cyclase/dehydrase and lipid transport superfamily protein
## 17817 Kinase interacting (KIP1-like) family protein
## 17909 BTB and TAZ domain protein 1
## 19701
## 19024 bZIP transcription factor family protein
## 17435 LOB domain-containing protein 37
## 22519 Granulin repeat cysteine protease family protein
## 1820 NAD(P)-binding Rossmann-fold superfamily protein
## 22251 sulfur E2
## 14147 Protein of unknown function (DUF581)
## 27313 glutamine synthetase 1;4
GO-term enrichment analysis
Getting the gene names are informative but it is hard to determine whether a speicifc category of terms are over-represented in this analysis. So we will perform a GO-term enrichment analysis of all differentially expressed genes using a Hypergeometric test to determine if there is any GO-term that is enriched in this list.
To run the test using the GOstats package, we must first create a GOHyperGParams object as shown below. Here we will provided the genes in the list, the universe of all genes, which GOterm gene space we are interested in, a p-value cutoff, and if we want an additional conditional test. The conditional test looks at parent and child relationship of the GO-terms. If the parent term and the child term are over-represented, the parent-term has to be significantly more over-represented to show up in the list of over-represented GO-terms. This avoids the problem where many over-represented GO-terms are parent or child of another and the terms appear to be very redundant.
library(GOstats)
## Loading required package: Category
## Loading required package: Matrix
## Warning: package 'Matrix' was built under R version 3.2.5
##
## Attaching package: 'Matrix'
## The following object is masked from 'package:IRanges':
##
## expand
## Loading required package: GO.db
## Loading required package: DBI
## Warning: package 'DBI' was built under R version 3.2.5
##
## Loading required package: graph
##
## Attaching package: 'graph'
## The following object is masked from 'package:Biostrings':
##
## complement
##
## Attaching package: 'GOstats'
## The following object is masked from 'package:AnnotationDbi':
##
## makeGOGraph
library(GO.db) library(Category) library(org.At.tair.db)
##
params=new("GOHyperGParams",
geneIds=rownames(resSig),
universeGeneIds=rownames(counts_filtered),
annotation="org.At.tair",
ontology="BP",
pvalueCutoff=0.001,
conditional=TRUE,
testDirection="over")
To get actually perform the test we use the hyperGTest function and to see the results simply use the summary function.
(overRepresented=hyperGTest(params))
## Gene to GO BP Conditional test for over-representation
## 827 GO BP ids tested (29 have p < 0.001)
## Selected gene set size: 76
## Gene universe size: 5211
## Annotation package: org.At.tair
summary(overRepresented)[,c(1,2,5,6,7)]
## GOBPID Pvalue Count Size
## 1 GO:0015706 4.050961e-09 14 132
## 2 GO:0010167 5.451615e-09 14 135
## 3 GO:0006820 7.463873e-08 18 284
## 4 GO:0019362 2.034569e-06 14 216
## 5 GO:0019676 2.982589e-06 3 3
## 6 GO:0042221 3.334282e-06 44 1684
## 7 GO:0006081 2.818555e-05 9 111
## 8 GO:0006098 4.647875e-05 6 46
## 9 GO:0006732 8.676677e-05 14 300
## 10 GO:0034284 1.178504e-04 8 104
## 11 GO:0006096 2.066368e-04 10 176
## 12 GO:0006165 2.066368e-04 10 176
## 13 GO:0009135 2.066368e-04 10 176
## 14 GO:0009185 2.066368e-04 10 176
## 15 GO:0046031 2.066368e-04 10 176
## 16 GO:0043648 2.072004e-04 4 21
## 17 GO:0009750 3.010900e-04 7 90
## 18 GO:0042128 3.324302e-04 3 10
## 19 GO:0043436 4.005847e-04 24 821
## 20 GO:0009144 4.180460e-04 10 192
## 21 GO:0009199 4.180460e-04 10 192
## 22 GO:0046034 4.180460e-04 10 192
## 23 GO:0016052 4.479565e-04 12 269
## 24 GO:0009126 5.129576e-04 10 197
## 25 GO:0009161 5.556402e-04 10 199
## 26 GO:0042278 7.017889e-04 10 205
## 27 GO:0009119 7.570648e-04 10 207
## 28 GO:0046686 8.329956e-04 14 373
## 29 GO:2001057 9.668296e-04 3 14
## Term
## 1 nitrate transport
## 2 response to nitrate
## 3 anion transport
## 4 pyridine nucleotide metabolic process
## 5 ammonia assimilation cycle
## 6 response to chemical
## 7 cellular aldehyde metabolic process
## 8 pentose-phosphate shunt
## 9 coenzyme metabolic process
## 10 response to monosaccharide
## 11 glycolytic process
## 12 nucleoside diphosphate phosphorylation
## 13 purine nucleoside diphosphate metabolic process
## 14 ribonucleoside diphosphate metabolic process
## 15 ADP metabolic process
## 16 dicarboxylic acid metabolic process
## 17 response to fructose
## 18 nitrate assimilation
## 19 oxoacid metabolic process
## 20 purine nucleoside triphosphate metabolic process
## 21 ribonucleoside triphosphate metabolic process
## 22 ATP metabolic process
## 23 carbohydrate catabolic process
## 24 purine nucleoside monophosphate metabolic process
## 25 ribonucleoside monophosphate metabolic process
## 26 purine nucleoside metabolic process
## 27 ribonucleoside metabolic process
## 28 response to cadmium ion
## 29 reactive nitrogen species metabolic process
Creating Heatmaps
Clustering is a powerful way of grouping together genes based on similarity of their expression profiles. Here we
will use an enhanced version of the heatmap function that has built-in clustering method.
To use heatmap.2 we need to install gplots.
Please note that gplots is not a part of Bioconductor so you have to install it the standard way by using
install.packages("gplots")
library("gplots")
## Attaching package: ‘gplots’
##
## The following object is masked from ‘package:IRanges’:
##
## space
##
## The following object is masked from ‘package:S4Vectors’:
##
## space
##
## The following object is masked from ‘package:stats’:
##
## lowess
##
By default the built-in clustering method uses Euclidean distance to determine similarity of genes instead of pearson correlation and uses the complete linkage method instead of average. Luckily heatmap.2 allows us to redefine the distance and hclust functions by providing our own. Below are two functions that allow us to make the adjustment.
hclust2 <- function(x, method="average", ...) {
hclust(x, method=method, ...)
}
dist2 <- function(x, ...) {
as.dist(1-cor(t(x), method="pearson"))
}
DESeq also provides a normalized version of the counts data which will become useful when performing advanced transcriptome analysis such as clustering. So let’s retrieve the normalized values and then create a new dataframe with expression values of just the differentially expressed genes.
resSig = c(rownames(resSigind),rownames(resSigrep)) normalized = rlog(counts_filtered, blind=FALSE) rownames(normalized)=rownames(counts_filtered) sigGenes.normalized = normalized[resSig,]
Now we will run the heatmap.2 function to create heatmap of the differentially expressed genes.
heatmap.2(as.matrix(sigGenes.normalized),
col=topo.colors(75),
hclustfun=hclust2,
distfun=dist2,
scale="row",
cexCol=0.6,
Colv=TRUE,
sepcolor="black",
dendrogram="both",
key=TRUE,
symkey=FALSE,
density.info="none",
trace="none",
cexRow=0.4)
The heatmap shows two distint groups, a larger one going up, and a smaller one going down.
#Clustering
Now id we wanted to get the gene names for the two clusters we can perform the clustering on our own and save them in different vectors.
sighclust=hclust(as.dist(1-cor(t(sigGenes.normalized))), method="average") sighclustgroups = cutree(sighclust, k=2) sighclust_g1= names(which(sighclustgroups==1)) sighclust_g2= names(which(sighclustgroups==2))
The hclust function is performing the heirarchichal clustering. Note that we have to first transpose the normalized matrix before we perform the correlation since it does this column by column. We then convert the correlation value to a distance measure by subtracting it from 1 and finally we tell R to make it into a distance object.
cutree is deciding where to cut the tree. We can use the k option, which allows you to specific the total number of final groups you want or the h option which allows you to pick a distance cutoff. The result is simply a vector of group identifiers (either 1 or 2) and then we simply identify which genes are in which cluster group.
Now you can do your Go-term enrichment on each group !