Metagenomics is the study of genetic material recovered directly from environmental samples. This is useful when attempting to understand what microbes are present and what they are doing in a particular environment.
Some common examples of sample sites are:



Why Metagenomics?
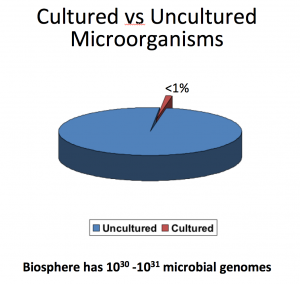
Conventional methods of studying microbiomes are extremely lossy – the vast majority of microbes cannot be cultured by plating due to many factors that we are not quite aware of.
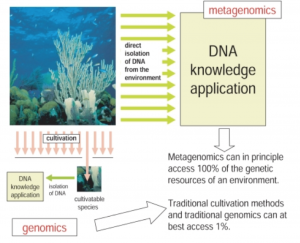
The traditional workflow consisted of collecting the sample, plating it, and sequencing what grows – the main limitation being you can only sequence what grows. Current technology allows us to completely bypass the culturing step and instead extract nucleic acids directly from a sample, granting access to theoretically 100% of the genetic content in the sample.
With this new methodology, the entire microbial population can be considered for the following questions, which are asked in every metagenomics project:
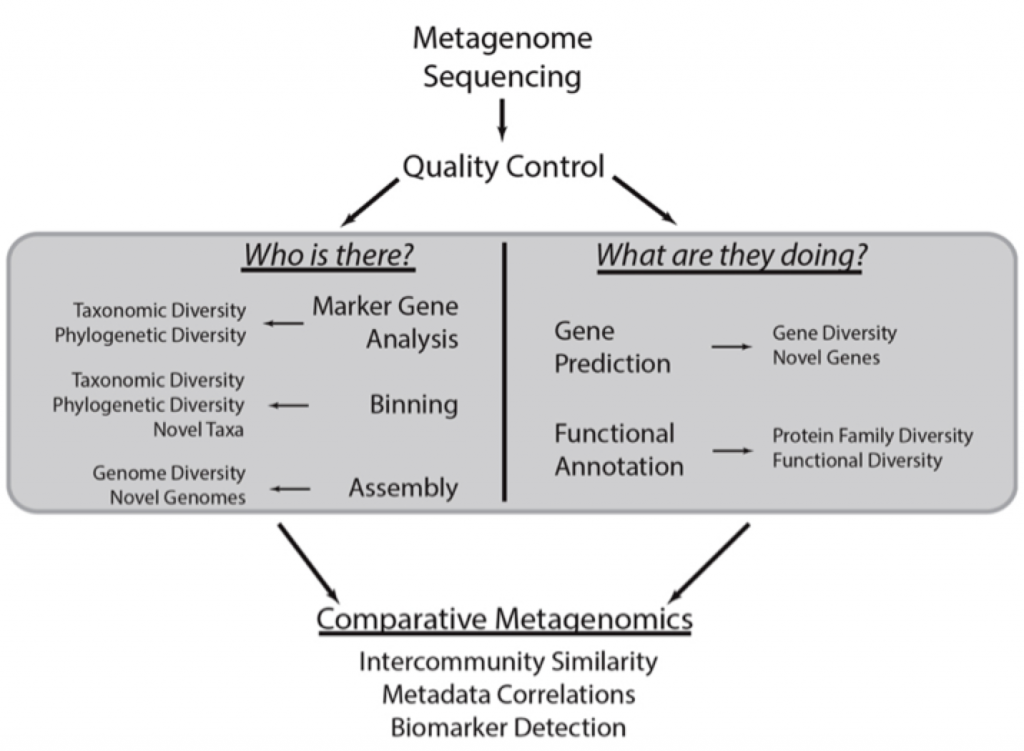
Being able to answer these questions enables us to define a ‘healthy’ microbiome that we can then use to treat disease, better protect and grow crops, clean up oil spills, and many other applications.
Types of Metagenomics
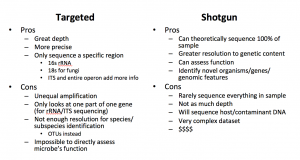
There are two types of metagenomics approaches: Targeted sequencing and metagenomic shotgun sequencing. The former is becoming phased out as sequencing costs fall and the technology improves but it is still used frequently as each approach has their own pros and cons. However, both can answer the question of who is in your sample, but only shotgun metagenomics can truly address their function.
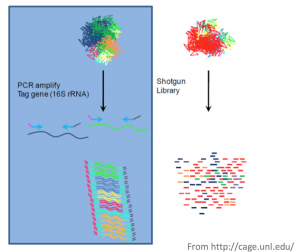
Targeted Metagenomics
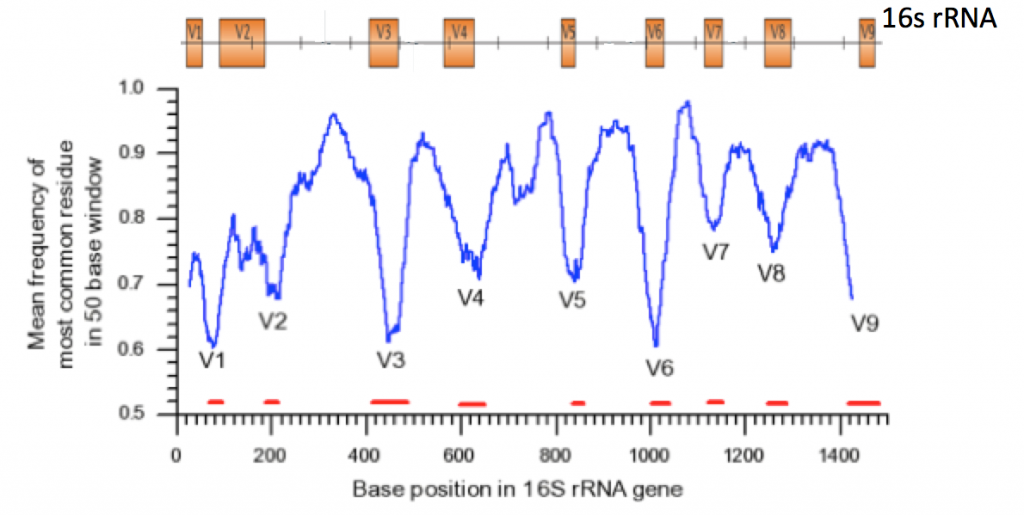
In this application, certain conserved regions (16s rRNA, 18s rRNA, ITS regions) are amplified with PCR primers and sequenced. These conserved regions have variable regions that allow for identification of different groups of organisms. However, identifying these organisms down to the species level is near impossible. Another drawback is that there is no way to directly assess function from these data.
Shotgun Metagenomics
This method is non-discriminant in that it will sequence everything in your sample. This not only can assign taxonomy and quantify the organisms down to the species level, but it can also assign function as it will sequence many different parts of each organism’s genome. If your data are good enough you can reconstruct entire genomes, genes, as well as infer different pathways.
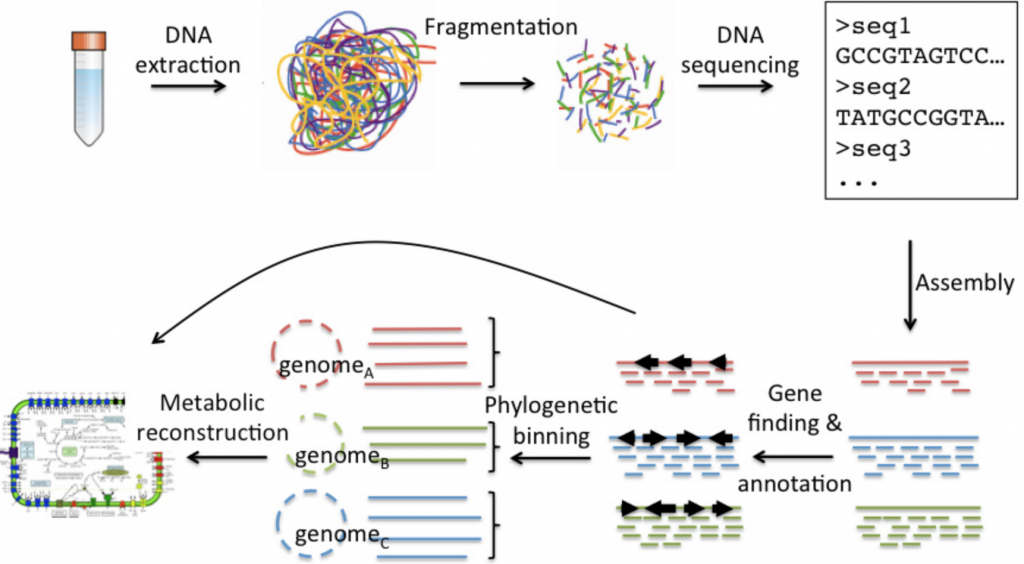
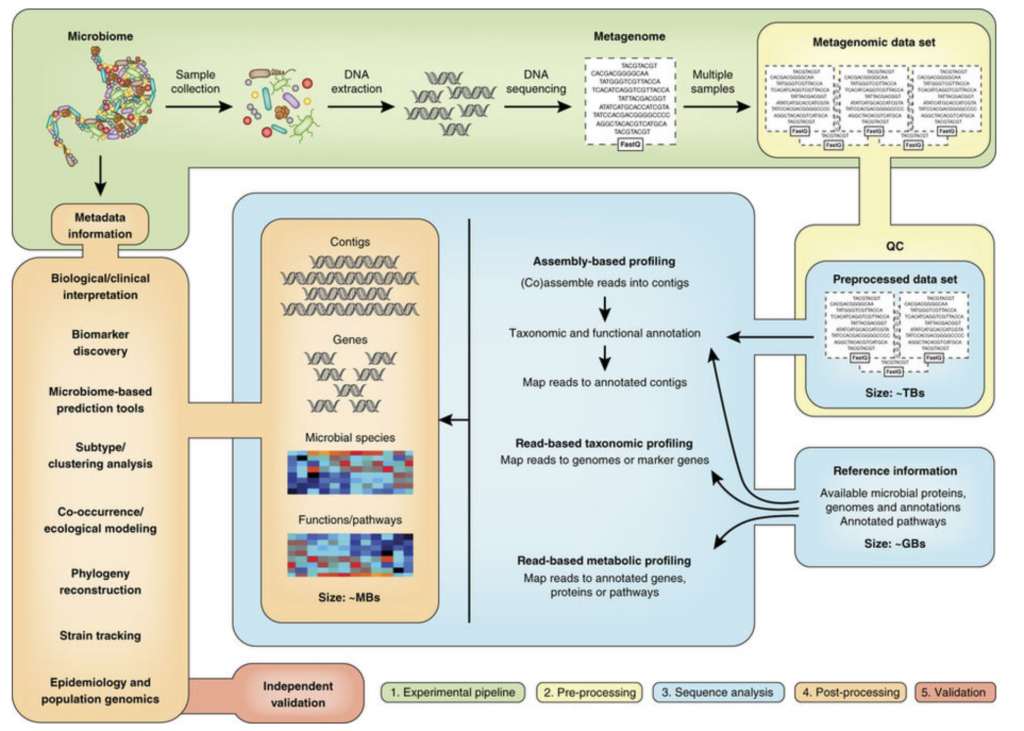
Metagenomic Study Design
Below is an overview of metagenomics analysis pipelines. Note that nearly all questions asked by targeted sequencing can be addressed with shotgun metagenomics so this design can be applies to both methods.
You can view the slides from my 2017 presentation here.