Introduction
RNA-seq is essentially the sequence of RNA molecules from either a specific cell, tissue, or species. There are two main motivations for sequencing RNA:
- Identifying differential expression of genes by comparing different samples.
- Attempt to capture all RNA molecules in a given species.
In the case where a species does not have a sequenced genome, the researcher has to do (2) before they can do (1).
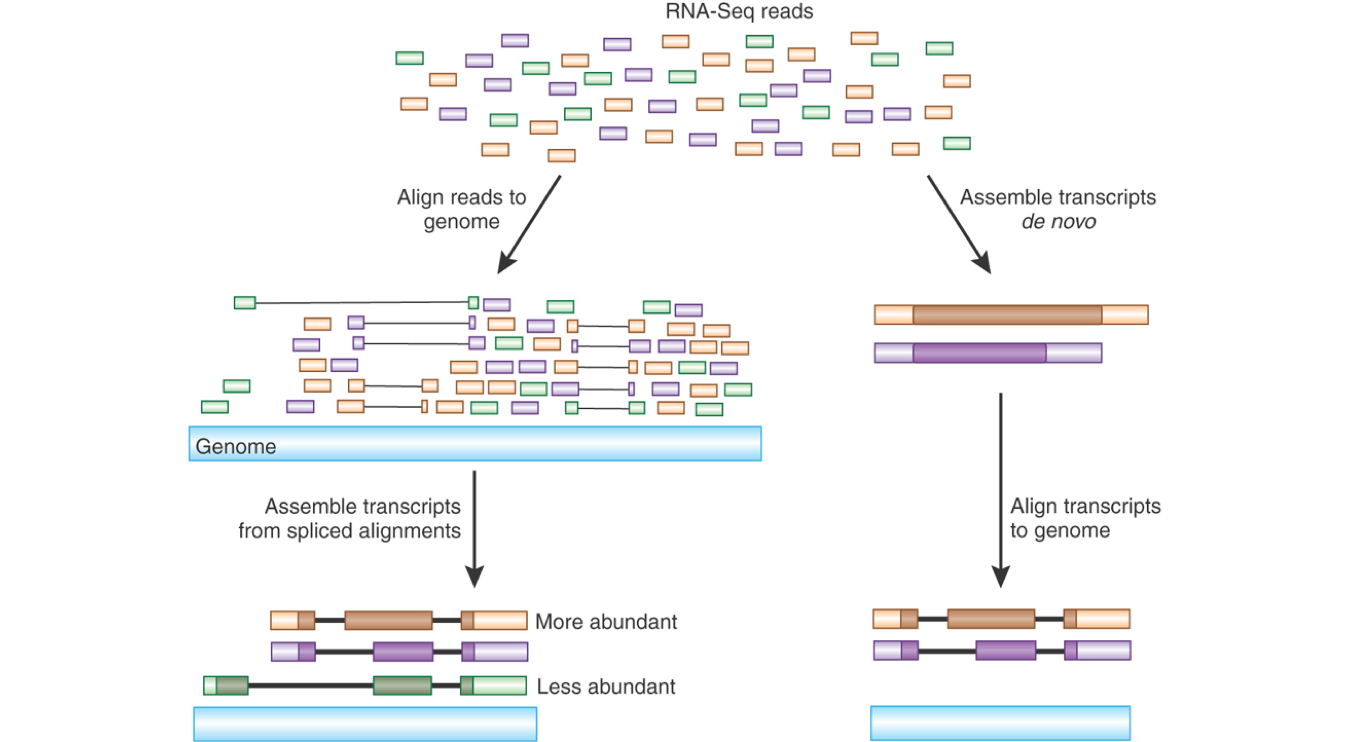
(Hass and Zody, Advancing RNA-Seq analysis, Nature Biotechnology 28:421-423)
In addition to RNA sequencing, Microarray technologies are an alternative method to determining differentially expressed genes. The main limitations of Microarray technologies are:
- Knowledge of reference genome is required to determine unique probes
- Can only determine differentially expressed for genes that are represented on the chip
- GeneChips do not have enough resolution to differentiate differential expression from different isoforms of the same gene.
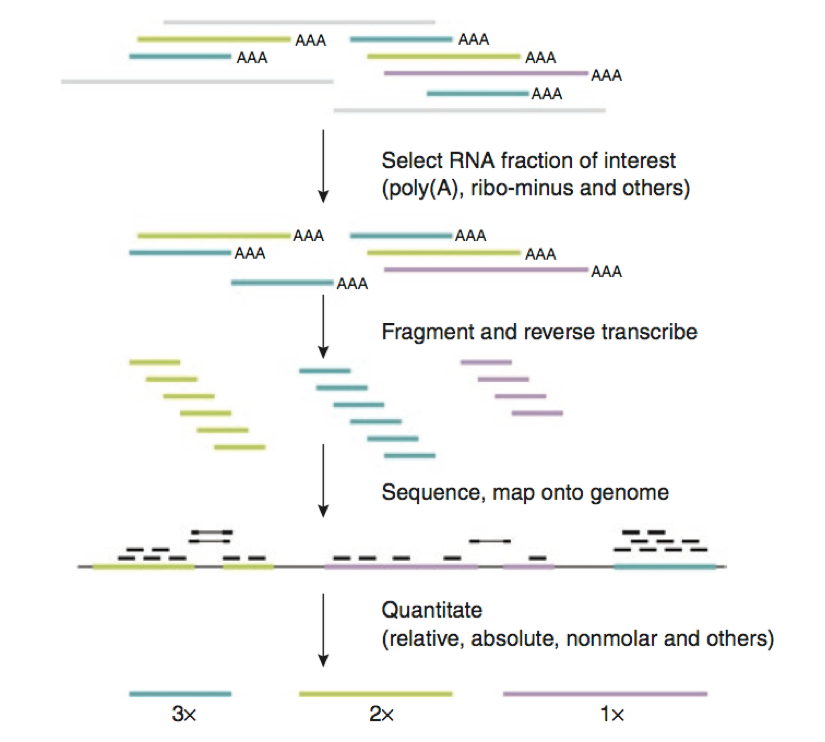
(Pepke S., Wold B., Mortazavi A., Computation for ChIP-seq and RNA-seq studies. Nature Methods 6:11, Nov 2009)
There are several different method for normalization and differentially expressed genes. Wang et al (Nature Biotechnology 2014) compared results from Microarray and RNA-seq using different methods and their results show that genes with a higher level of expression are detected by most methods.
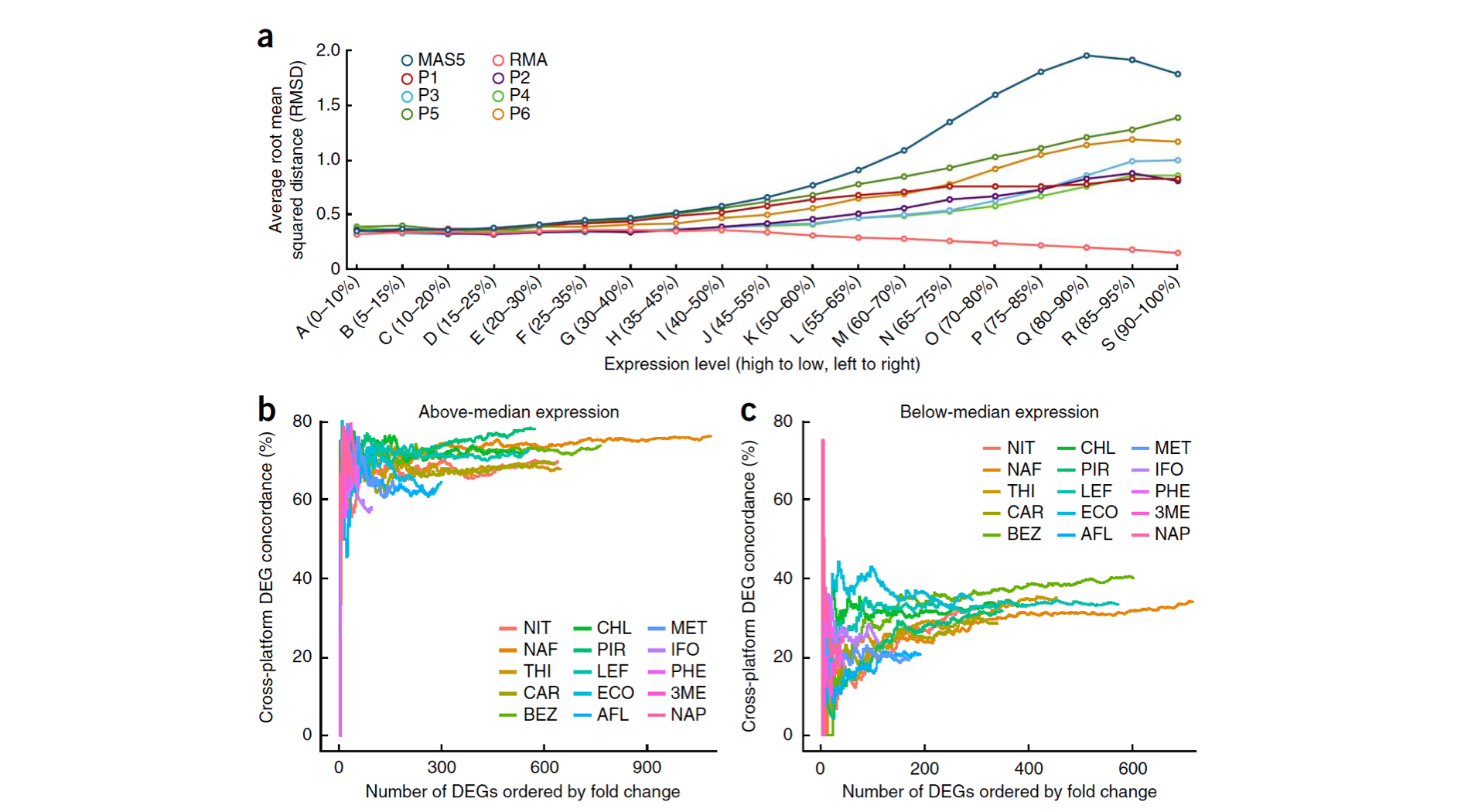
The workflow
- Quality Contol : Fastqc
-
Quality Trimming: Trimmomatic
-
Alignment: Tophat2 & Bowtie2
-
Index and Dedup: Picard
DESeq Example
Required R/Bioconductor packages
- Rsamtools
- GenomicFeatures
- GenomicAlignments
- DESeq
Dataset to download
https://drive.google.com/drive/folders/0B172nc4dAaaObEhfZkVxaUFLY28?usp=sharing