An in depth writeup about quality scores can be found here.
Quality scores are a way to assign confidence to a particular base within a read. Some sequencers have their own proprietary quality encoding but most have adopted Phred-33 encoding. Each quality score represents the probability of an incorrect basecall at that position.
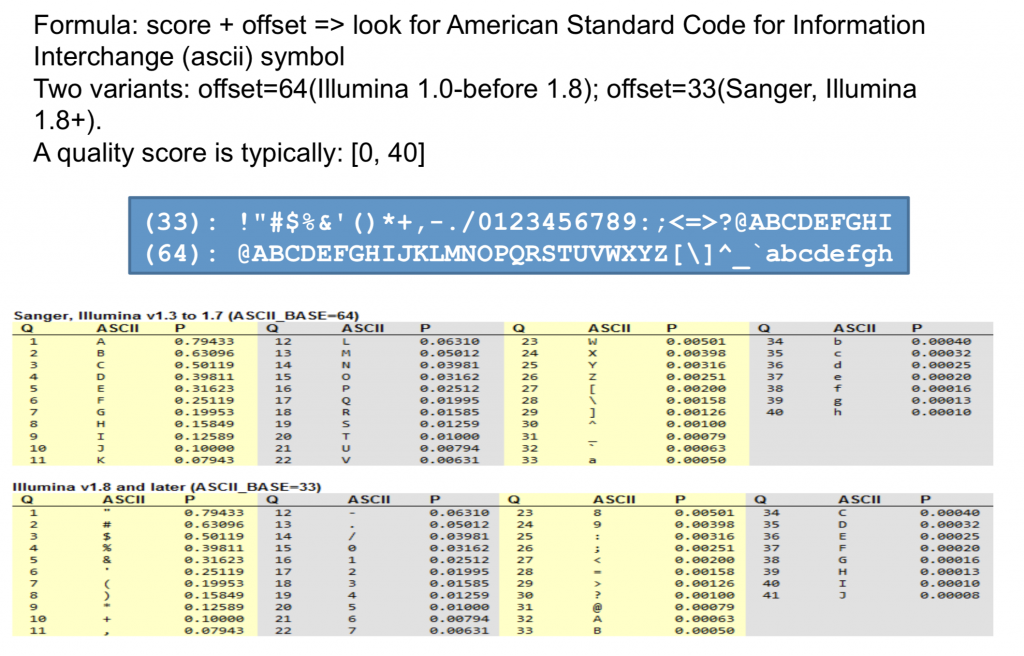
Phred Quality Score Encoding
Quality scores started as numbers (0-40) but have since changed to an ASCII encoding to reduce filesize and make working with this format a bit easier, however they still hold the same information. ASCII codes are assigned based on the formula found below. This table can serve as a lookup as you progress through your analysis.
Note that Phred-64 was only ever used by Illumina and is not deprecated.
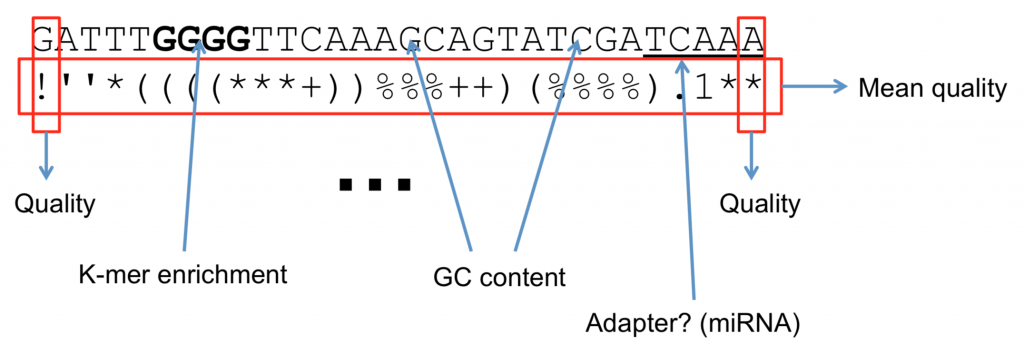
Quality Score Interpretation
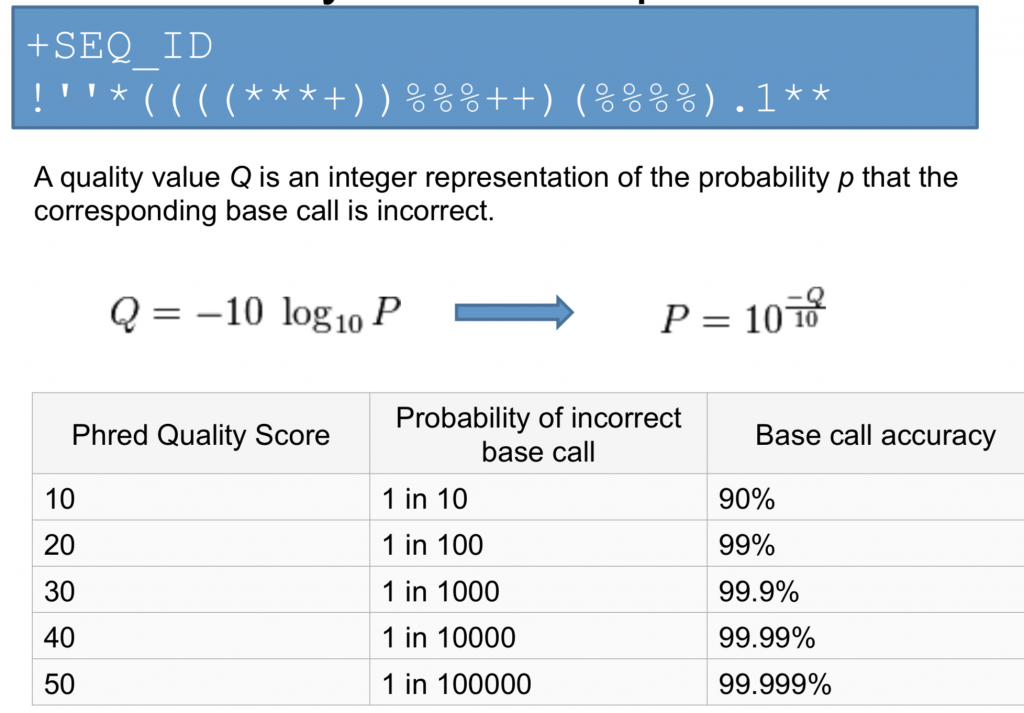
Once you know what each quality score represents you can then use this chart to understand the confidence in a particular base.
What are Quality Scores Good for?
As we mentioned earlier, many programs require the FastQ format, implying that they will use the quality score in a particular part of the analysis. Common uses are to filter bases or entire reads if a particular quality threshold isn’t met. An example of a threshold is the mean quality score for the read. That is: what’s the average score of all bases for an individual read? If the average Phred quality score is 10, what does that mean? Is this good enough to do SNP analysis?
What Software use Quality Scores?
The main purpose for these scores is to further provide evidence that the sequence, alignment, assembly, SNP are in fact real and not due to a problem in generating the sequences.
- Almost every QC software package use these.
- Variant detection/SNP calling algorithms
- Assemblers
- Aligners