Many different companies have their own sequencing technologies, some better suited for certain applications than others, but the concepts boil down to the same concept: identify each nucleotide in a particular molecule.
The Genomics Core here at NYU New York City and in Abu Dhabi have a few different sequencers to choose from.

There are three main steps in NGS:
- Sample collection/preparation
- Amplification
- Basecalling
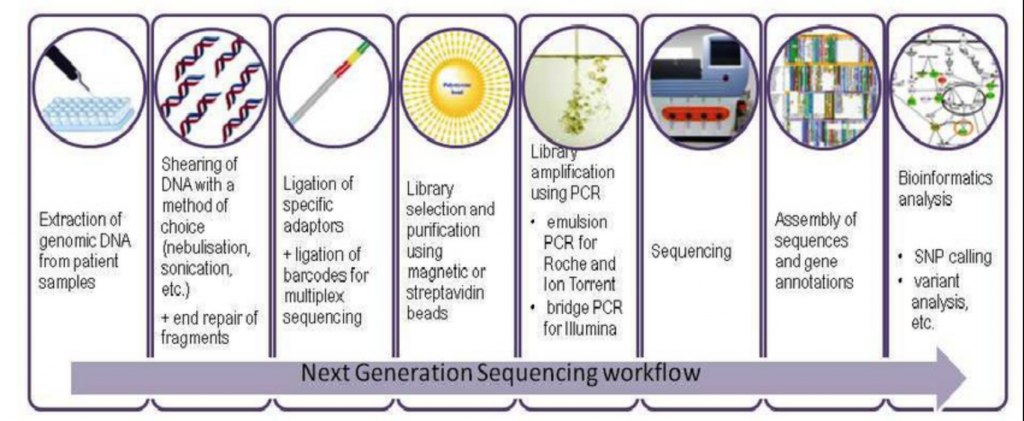
Sample Collection and Preparation
This step involves gathering the nucleic acids from your organism of interest. The collection method differs based on what the sample is, but the preparation usually involves isolating and purifying the nucleic acids, shearing your nucleic acids to a certain size, amplification of your product, and ligation of sequencing adaptors (small fragments of DNA used to anchor the molecule of interest onto the flowcell).
Single-end vs Paired-end Sequencing
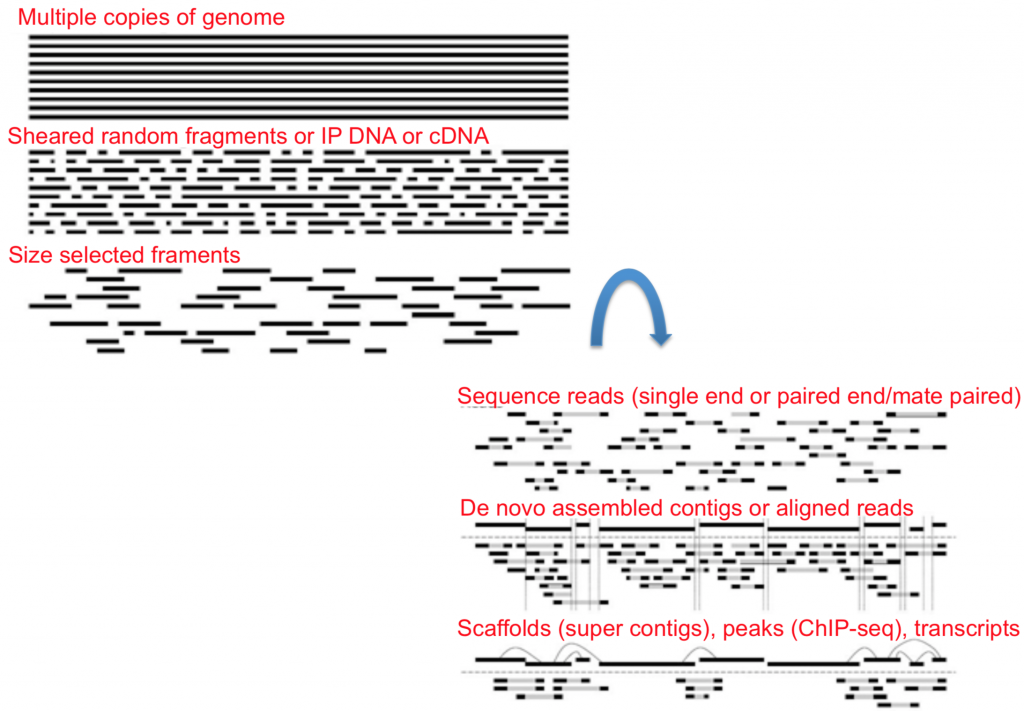
Once you have your nucleic acids ready to go you can then choose whether you want single-end or paired-end data.


To summarize, the illustration below shows each step of the library prep once nucleic acids are isolated and amplified.
Sequencing
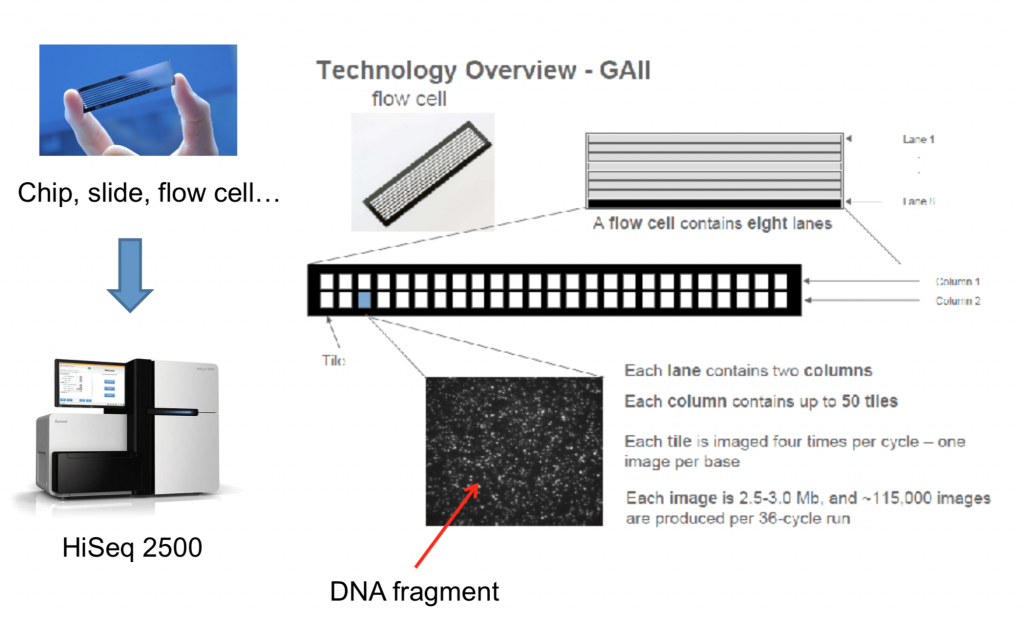
To better understand how sequencing is done on the machine, the let’s look over the diagram below. This shows the physical layout of the flowcell of which the DNA is loaded onto. You may hear some labs say “we have two lanes on a HiSeq”. This means that their sample was loaded onto two lanes, while some other lab(s) utilized the rest. This is a way to drive the cost down and avoid overkill.
Amplification and basecalling are two essential steps in the Illumina sequencing technology. In the video below you can see how this works (the visuals help!).